Food borne diseases are increasingly recognized as a major public health problem both nationally as well as globally. Despite major advances and improvements in hygiene, quality of food, water, sanitation and detection of food borne pathogens, food borne diseases remain one of the leading public health concerns. There is tremendous economic burden of treating animals and human patients affected by antimicrobial resistant pathogens and critical knowledge gaps exist in the complex chain of events that leads to dissemination and persistence of these bacterial pathogens. There is no clear consensus on the significance of ecological and management factors in the dissemination of such antimicrobial resistant strains in humans. In addition, characterizing the pathogen to specific strain level is time consuming.
Research in our lab is focused on understanding the molecular epidemiology of important multidrug resistant (MDR) food borne pathogens at the pre-harvest and post-harvest food safety levels. Our specific goal is to determine the dynamics of Salmonella, Campylobacter and Clostridium in food animals, retail meat, humans and the environment. With this information, we plan to achieve our long term goal of reducing the burden of infections caused by bacterial pathogens in food animals and humans by:
- Determining the risk factors and understanding the dissemination of antimicrobial resistant strains in animals, humans and their environment,
- Characterizing and elucidating the mechanisms of antimicrobial resistance at the molecular level,
- Understanding the genotypic diversity and population dynamics of these bacterial populations through phylogenetic analysis,
- Developing diagnostic methods that will aid in the rapid identification and characterization of bacterial pathogens and,
- Conveying the results of these studies to the stakeholders and developing new curricula aimed at providing education and training of veterinary, animal and food science students.
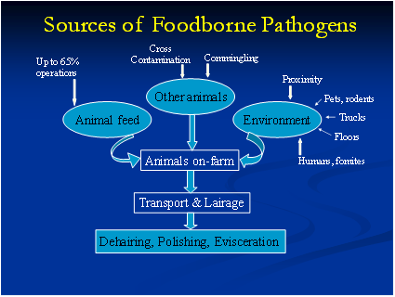
The underlying rationale behind this objective is that it is important to determine the risk factors that aid in the dissemination and persistence of important bacterial pathogens at the pre and post harvest food safety levels. It is imperative that we pin point the exact sources that transmit these pathogens to both pigs and humans. Once we determine these foci, we can devise appropriate strategies to eliminate them, thereby, reducing the burden of infection and illness in the host.
To determine these sources and risk factors, we conduct field sampling that includes sample collection from the swine at farm, slaughter and their environment. Sampling is also done at the retail level as part of our post-harvest testing. We isolate Salmonella, Campylobacter and Clostridium difficile from these samples and further characterize them at the phenotypic and genotypic levels. In addition, appropriate data is collected on every trip via questionnaires. Finally, we conduct risk factors studies using the data collected through these questionnaires after employing appropriate statistical methods.
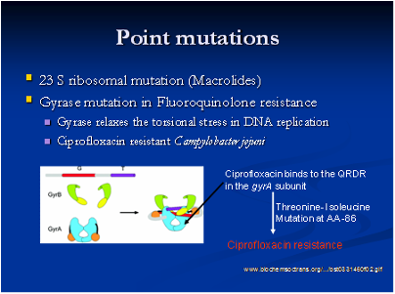
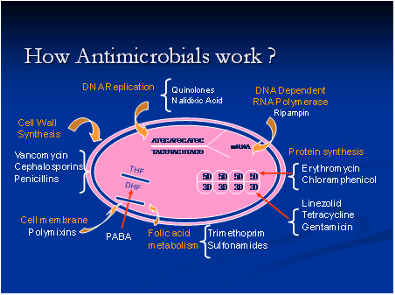
The emergence and spread of multi drug resistant bacterial strains has become a major concern among public health officials and the general public. There is a tremendous burden of infections caused by antimicrobial resistant bacterial strains in both humans and animals. Bacterial pathogens become resistant to different antimicrobials through a number of mechanisms, including enzymatic degradation of the antimicrobial, mutation in the antimicrobial target site, active efflux of the antimicrobial across the cell membrane, decreases in the cell wall permeability to antimicrobials, and the development of alternate metabolic pathways to circumvent antimicrobial interference. Therefore, it is important to determine the mechanisms and resistance determinants that confer resistance in these bacterial strains. We also want to understand the signal machinery involved in the entire process of antimicrobial resistance activation in these pathogens.
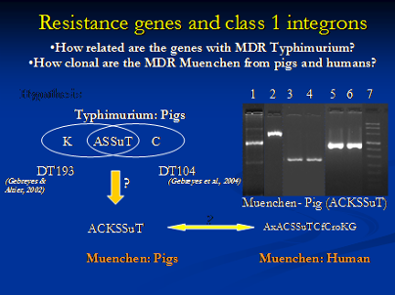
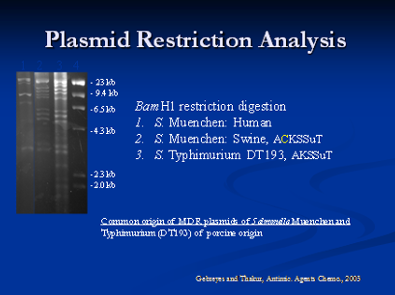
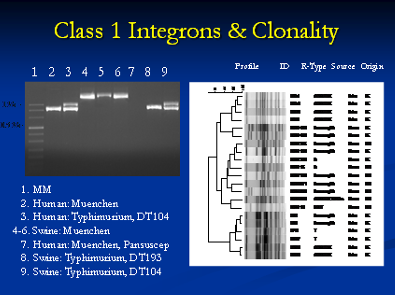
Molecular characterization of antimicrobial resistance is very important as it helps to determine the resistance genes, their location either on a plasmid or chromosome and other novel resistance mechanisms that might be present. For example, we have characterized Class-I integrons in different Salmonella serovars and identified evidence of resistance gene transfer between different serotypes of Salmonella in swine and humans. In addition, we have characterized chromosomal mutations responsible for fluoroquinolone and erythromycin resistance in Campylobacter isolated from swine. A plasmid, or integron, which has acquired the genetic determinants for multiple genes, can transmit multiple antimicrobial resistance determinants in a single exchange event. Plasmid analysis is also an integral component of this part of our research. We have characterized the exchange of plasmids between different serovars of Salmonella in swine.
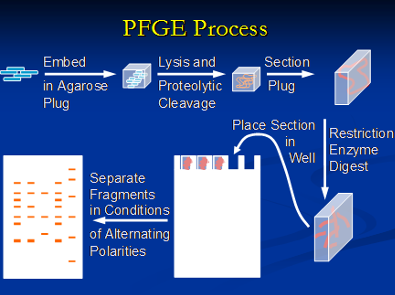
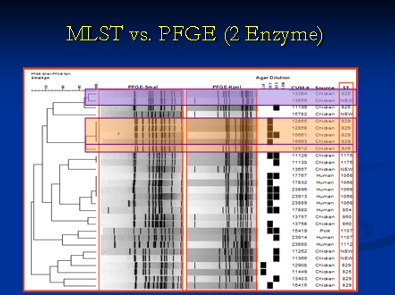
It is important to understand the relationship between different bacterial isolates and strains at the genotypic level. This information is very powerful and aids in source determination and outbreak investigations. We utilize different genotypic tools for this purpose including Pulsed field gel electrophoresis (PFGE), Multi locus sequence typing (MLST), Amplified fragment length polymorphism (AFLP) and Ribotyping to name a few. PFGE has been used by PulseNet within the United States for nationwide surveillance of many important food borne pathogens. The basic principle of PFGE involves purifying the pathogen genome which is embedded in agarose plugs followed by cutting of the genome at specific sites using restriction enzymes. Selection of the appropriate restriction enzyme, preferentially a rare-cutter, depends on the genome sequence of the pathogen and the particular nucleotide sequence length that the restriction enzyme recognizes and cleaves.
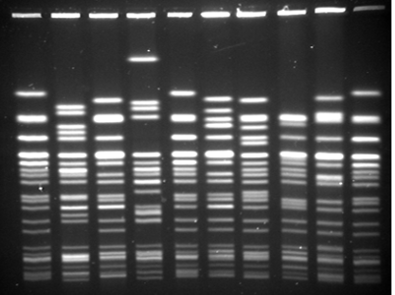
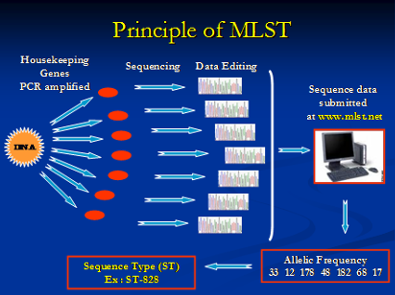
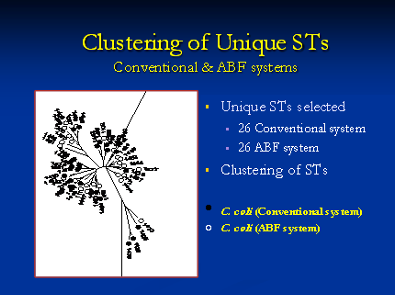
MLST is a suitable technique for genotyping bacterial populations that are weakly clonal. This method indexes the neutral variation seen in the housekeeping genes which evolve slowly and are stabilized for conserving the important metabolic proteins that they code. This is especially important to consider in pathogens that have a hypervariable genome which results due to the spontaneous intramolecular genomic recombinations. Briefly, all the housekeeping genes are amplified using PCR followed by sequencing of the amplified product. An allelic profile is generated that leads to the generation of a sequence type (ST). Data generated by this method is accurate since it involves comparing nucleotides of specific sequence and size. The ST information can be used to generate a dendrogram with isolates sharing similar genotypes are grouped together. The data helps us to generate dendrograms as shown below that is further utilized to determine if a bacterial population is clonal or diverse in its genotype.
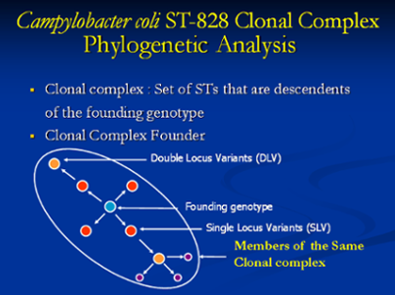
A major focus of our research is to understand the evolutionary relationship bacterial isolates of a species share with each other. This analysis helps in understanding the global epidemiology of bacterial pathogens. It also has a role in outbreak investigations and helps to understand the epidemiology of sporadic cases acquired from various sources. These evolutionary relationships can be generated based on the association of antimicrobial resistance or virulence gene profile with the source or geographic location of these isolates.
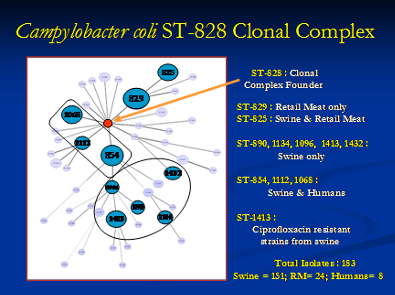
This is all the more important when we compare isolates from different hosts including humans at the phylogenetic level. We utilize DNA sequence data of the different housekeeping genes generated when the bacterial isolates are genotyped by MLST. We have identified unique Campylobacter coli Sequence Types (ST) in different pig populations at farm and slaughter indicating niche adaptation. Even though we identified genotypically diverse Campylobacter coli populations from humans and retail meats, they were grouped under the same clonal complex indicating common ancestry. The detection of specific/diverse lineages in different hosts will aid us in comparing isolates from these sources around the world and be useful in understanding the global epidemiology of pathogen.
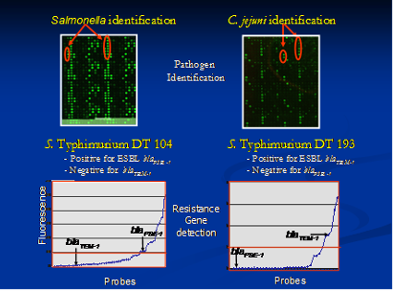
Rapid and sensitive detection of pathogens including characterizing the antimicrobial and virulence gene profile is of prime importance. Detection and characterization of these pathogens during outbreak scenarios usually takes a lot of time. Our aim is to develop rapid and sensitive DNA based methods that will help in both detection and quantification of important food borne bacterial pathogens. We are utilizing different methods including real time PCR, pyrosequencing, sequencing and microarrays for developing these tests.
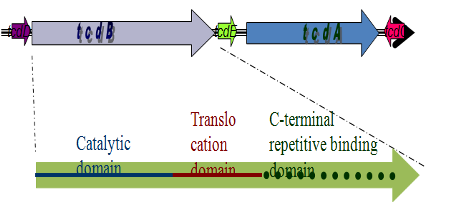
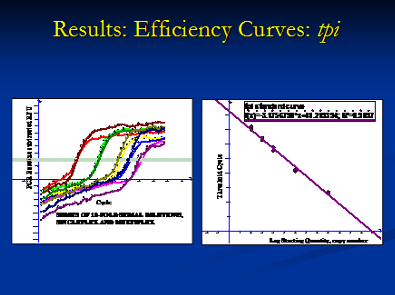
Quantitative RT-PCR can be an effective tool for studying food borne pathogens, because it can be used to rapidly identify and quantify multiple characters simultaneously. Using probes that fluoresce when incorporated into a DNA replication, we can estimate the starting number of DNA target copies. We have recently developed a quantitative multiplex assay and characterized the pathogenicity of Clostridium, a microbe which has recently emerged as a public health threat and potential food borne pathogen. We aim to quantify pathogenic and non-pathogenic Clostridium in various fecal and retail meat samples to better understand the epidemiological pattern of the pathogen. Assessing the quantity and pathogenicity at these steps may illuminate how this organism is disseminated. Our multiplex reactions were able to correctly characterize the pathogenicity of our samples 100% of the time, for all potential profiles. Our sensitivity, specificity, and predictive values were also 100%.
We are currently collaborating to develop an oligonucleotide based microarray chip for the simultaneous identification and characterization of virulence and antimicrobial resistance genes present in bacterial and viral food borne pathogens. The main aim of the proposed research program is to develop DNA based microarray for rapid, sensitive and definitive detection, diagnosis and characterization of the most important infectious bacterial (Campylobacter, Salmonella and Yersinia) and viral (Noroviruses) pathogens. We have identified specific target regions and designed probes that are specific to the pathogen of interest. The fully developed chip will increase the capacity of rapid identification and characterization of the food borne pathogen. The chip will greatly reduce the time taken and cost of detecting a pathogen, especially in outbreak scenarios. In addition, it will provide a sensitive and specific method to screen samples for resistance, virulence genes, and zoonotic pathogens of consequence to human health.